This web page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison
LDLR Protein Phylogeny
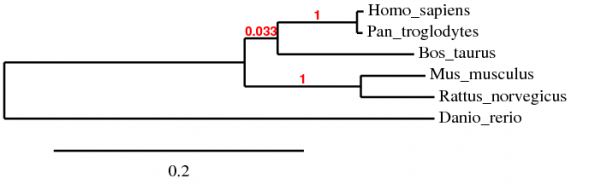
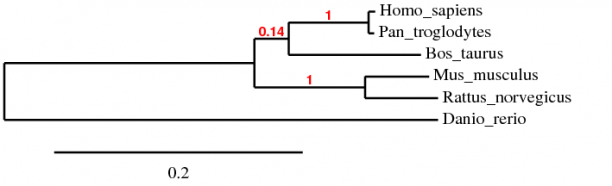
Protein phylogeny was also analyzed with phylogeny.fr using the amino acid sequences of all the homologs. Again the MUSCLE algorithm was used, along with TreeDyn in order to create the evolutionary tree. Because the amino acid sequences were within the limits of the ClustalW algorithm, a tree was also created in order to compare both.
Analysis
As with the gene phylogeny, the MUSCLE and ClustalW trees followed the sequence homology determined by BLAST. There is only a slight difference in the evolutionary distance of the divergence of Bos taurus, a common ancestor to Homo sapiens and Pan troglodytes. This can be attributed to the algorithmic differences of MUSCLE and ClustalW.