This web page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison
LDLR Protein Interactions
Protein interaction networks were built using the STRING and Osprey programs. STRING is available online, while Osprey is a program that needs to be downloaded. The results of LDLR interactions are found below.
STRING
The STRING database takes protein information from multiple databases and websites in order to create specific interaction webs. A more in depth explanation of how these interactions are found can be found here. In the below STRING figures, a default cutoff of 10 interactions was used. This allowed for a somewhat complex, yet manageable web that only included the most statistically significant interactions. The different colored lines represent how the proteins it connects have been shown to interact: yellow is through text; purple is experiments; turquoise is databases; light blue is homology. Also, I believe that some proteins are represented with small circles because they don't have 3-D structures associated with them yet. As can be seen within the figures, a 3-D structure can be seen in all of the larger proteins. Although I have not found an official explanation of the size differences, this seems to be reasonable.
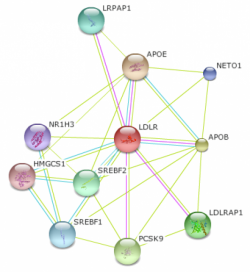
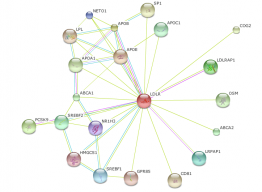
Figure 1-Homo sapiens LDLR interaction web. Click to enlarge.
Figure 1 shows the Homo sapiens LDLR interaction web with the default STRING settings. The program initially creates webs using a medium confidence level of 0.400 and only shows, at most ten interactions. This first web gave ten proteins, all having confidence scores above 0.960. With confidence scores that are so high it is reasonable to assume that all ten proteins interact with LDLR. The top three proteins (APOE, APOB, and PCSK9) all had confidence scores of 0.999. ApoB is the main apoprotein found on LDL particles and it is bound by LDLR which leads to internalization. A specific mutation within the APOB gene leads to a disease known as familial defective apob-100 (FDB). This disease is basically identical to FH: autosomal dominantly inherited, increased LDL levels and heart disease risk (1). LDLR is also able to bind ApoE, the main apoprotein of chylomicrons and chylomicron remnants (1). PCSK9, proprotein convertase subtilisin/kexin 9, is a more recently discovered protein to be involved in cholesterol metabolism. PCSK9 is a protease that is able to bind and degrade LDLR. This regulates LDL receptor levels in cells, in turn effecting cholesterol uptake. Mutations in PCSK9 have been shown to cause increased cholesterol resulting in an indistinguishable phenotype to FH (1, 2).
Because the initial interaction web proteins had such high confidence scores, I wanted to expand the web to include more significant proteins. By selecting the highest confidence cutoff (0.900) and allowing for a maximum of 50 protein interactions, a new web was created. Figure 2 shows this interaction web, which includes 20 proteins, and the lowest confidence score was 0.906. As expected, more proteins involved in cholesterol metabolism were found, including two apoproteins (APOA1 and APOC1) and LPL (lipoprotein lipase precursor). LPL hydrolyzes VLDL particles into VLDL remnants which are either cleared from the blood via LDLR or are further processed into LDL particles (1).
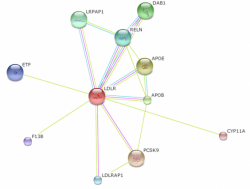
Figure 3 shows the interaction web for Pan troglodytes LDLR. This web, using the default settings, is less intricate and confident compared to homo sapiens. This is most likely a reflection of less research on chimpanzee cholesterol metabolism. Although the top three proteins are again APOE, APOB, and PCSK9, the confidence scores are much lower (0.719, 0.710, and 0.813 respectively).
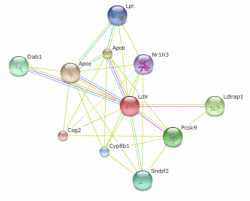
Figure 4 shows the interaction web for Mus musculus Ldlr. This web is much more similar to the Homo sapiens web compared to Pan troglodytes. Although chimpanzee is the closest relative to humans, it is not normally used as a model organism. Mouse, on the other hand, is the mammalian model organism of choice and this is reflected in the high confidence scores of these proteins.
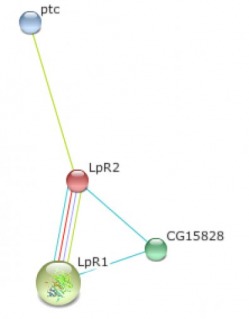
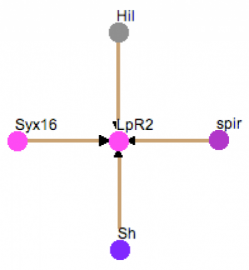
Finally, the Drosophila melanogaster protein LpR2 interaction web is also shown. Only three proteins were shown to interact with LpR2 and the confidence scores are fairly low. The small amount of information found on LpR2 is probably because of the simplistic nature of the organism. Fly cholesterol metabolism is not as complex as higher organisms and it also seems that little research has been done with this organism on cholesterol mechanisms.
Osprey
Osprey is a downloadable program that utilizes the Biological General Repository for Interaction Datasets (BioGRID) database. Using GO terms and information from primary literature Osprey creates protein interaction webs. You are able to select from multiple organism databases (budding yeast, mouse, fly, human, worm, fission yeast, rat, and zebra fish) when creating protein webs. The colors of the proteins in the web are determined by GO terms. I searched for interaction webs in each organism and only human and fly had protein interaction.
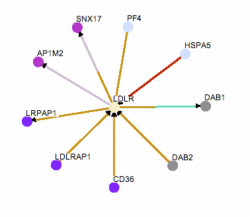
Figure 6-Homo sapiens LDLR interaction web. Click to enlarge.
The Homo sapiens interaction web is shown to the right. Osprey was able to find nine proteins that interact with LDLR. Most of the proteins are involved with protein transport and metabolism, however there is not as much lipid metabolism interaction compared to STRING. LRPAP1 and LDLRAP1 are the only two proteins found in both STRING and Osprey. LRPAP1, low density lipoprotein receptor-related protein associated protein 1, is a subunit of LDL receptor-related protein (LRP) which is also involved in cholesterol clearance (3). LDLRAP1, low density lipoprotein receptor adaptor protein 1, also known as autosomal recessive hypercholesterolemia (ARH) protein, mediates the interaction of LDLR and clathrin which aids in LDL internalization. Mutations in this gene can lead to an autosomal recessive disease that is phenotypically identical to FH (4).
Figure 7 gives the interaction web for Drosophila melanogaster. It is very simple, similar to the STRING web, probably because little research has been done on cholesterol metabolism in this organism. Syx16, spir, and Sh are all involved with protein transport, while Hil has no associated GO terms and has an unknown function. Syx16 is the only protein that has function somewhat related to LDLR; it is involved in vesicle docking and transport. Overall, I do not feel that this web is very helpful for studying LDLR and cholesterol metabolism. It is too simplistic and the proteins don't necessarily relate well to LDLR.
Analysis
Both STRING and Osprey are very simple programs that allow the easy creation of protein interaction webs. In general, I prefer STRING. The program has many more avaliable proteins and organisms, and the webs are more aesthetically pleasing compared to Osprey. Also, STRING allows expanding of webs by choosing confidence score cutoffs. Although you are able to choose the color of proteins in Osprey webs, they look less professional compared to STRING. I think that as the BioGRID database continues to grow, Osprey will become a better program.
The proteins found to be associated with LDLR and its homologs in STRING were more extensive and a better reflection of LDLR function compared to Osprey. Again this is because of a wider range of information available in STRING. Both programs found proteins associated with protein transport, which is easily related to LDLR function. Overall, protein interaction webs can provide an individual with a way to expand their research in a focused manner, similar to GO terms.
The proteins found to be associated with LDLR and its homologs in STRING were more extensive and a better reflection of LDLR function compared to Osprey. Again this is because of a wider range of information available in STRING. Both programs found proteins associated with protein transport, which is easily related to LDLR function. Overall, protein interaction webs can provide an individual with a way to expand their research in a focused manner, similar to GO terms.
BioCarta
BioCarta is a website that provides pre-made pathway visual aids. You can search for specific genes in order to find pathways that involve the protein.
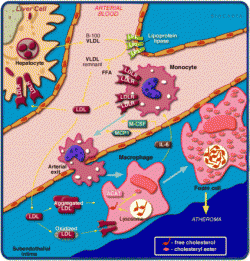
When I searched for LDLR, BioCarta returned four different pathways. The LDL pathway during atherosclerosis (Figure 8) was the most relevant result. An in depth explanation of the pathway can be viewed here. BioCarta provides a concrete visualization of protein interactions. By showing the actual cells and tissues where these proteins function, you are able to come full circle from initial findings in STRING and Osprey. Although not every protein found in STRING and Osprey are found in the BioCarta pathways, this program still helps understand the function of LDLR and its interacting proteins.
References
1. Rader, D. J., Cohen, J., & Hobbs, H. H. (2003). Monogenic hypercholesterolemia: new insights in pathogenesis and treatment. The Journal of Clinical Investigation, 111(12), 1795–1803. doi: 10.1172/JCI200318925
2. Mousavi, S. A., Berge, K. E., & Leren, T. P. (2009). The unique role of proprotein convertase subtilisin/kexin 9 in cholesterol homeostasis. Journal of Internal Medicine, 266(6), 507-19. doi: 10.1111/j.1365-2796.2009.02167.x
3. Pandey, P., Pradhan, S., & Mittal, B. (2008). LRP-associated protein gene (LRPAP1) and susceptibility to degenerative demetia. Genes, Brain and Behavior, 7(8), 943-50. doi: 10.1111/j.1601-183X.2008.00436.x
4. Soutar, A. K. (2010). Rare genetic causes of autosomal dominant or recessive hypercholesterolemia. IUBMB Life, 62(2), 125-31. doi: 10.1002/iub.299
2. Mousavi, S. A., Berge, K. E., & Leren, T. P. (2009). The unique role of proprotein convertase subtilisin/kexin 9 in cholesterol homeostasis. Journal of Internal Medicine, 266(6), 507-19. doi: 10.1111/j.1365-2796.2009.02167.x
3. Pandey, P., Pradhan, S., & Mittal, B. (2008). LRP-associated protein gene (LRPAP1) and susceptibility to degenerative demetia. Genes, Brain and Behavior, 7(8), 943-50. doi: 10.1111/j.1601-183X.2008.00436.x
4. Soutar, A. K. (2010). Rare genetic causes of autosomal dominant or recessive hypercholesterolemia. IUBMB Life, 62(2), 125-31. doi: 10.1002/iub.299