This web page was produced as an assignment for Genetics 677, an undergraduate course at UW-Madison
LDLR Protein Domains
In order to search for protein domains in the LDLR gene Pfam and SMART were used. All 860 amino acids were used for the query sequence in both programs. Similar domains were found in both programs, although the few differences will be highlighted below.
SMART Analysis
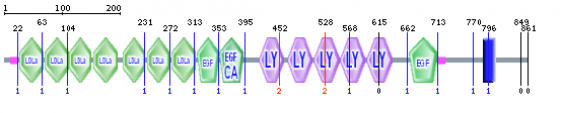
The above image shows the domains found within the LDLR protein using the normal SMART mode. As seen in figure 1, LDLR contains seven LDLa domains, three EGF/EGF-CA domains, five LY or LDLb domains, and a transmembrane domain. The transmembrane domain anchors LDLR into the plasma membrane, allowing the other domains to exist in the extracellular space (1, 2). The 3-D structures of the domains and biochemistry involved in LDL binding and release is explained below.
Pfam 3-D Domains
LDL Receptor Domain Class A (LDLa)
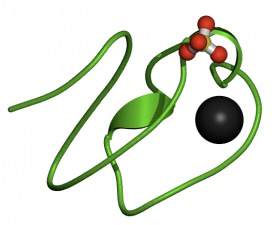
Figure 2-LDLa Domain. Click for website.
The LDLa domain is found seven times within the LDLR gene. The black area within in the domain is a calcium binding site. Calcium is needed for the domain to function properly. These cysteine rich domains are responsible for extracellular binding of LDL particles. Other negatively charged residues within these domains interact with positive residues on apoproteins during binding. Mutations that affect the binding properties of LDLa can drastically reduce LDL uptake and can cause FH (2).
Epidermal Growth Factor-like Domain (EGF)
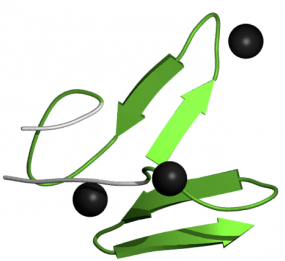
Figure 3-EGF Domain. Click for website.
EGF-like domains are found in many types of proteins and are conserved across multiple species. Proteins that contain this domain are often involved in protein-protein interactions, are receptors, or are growth factors. This indicates EGF-like domains are important for protein-ligand interactions, as with LDLR-LDL (3).
Calcium-binding EGF-like Domain (EGF-CA)

Figure 4-EGF-CA Domain
This domain is very similar to EGF-like domains, except that is has calcium binding capabilities. The proteins that have these EGF-like domains often need calcium in order to function correctly. Calcium binding occurs in the N-terminus of this domain (4). The image at right is actually depicting a pair of calcium-binding EGF-like domains interacting together.
LDL Receptor Repeat Class B (LDLb)
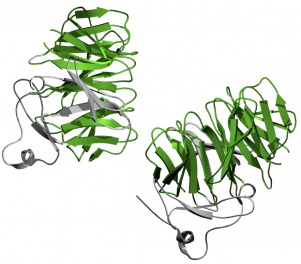
Figure 5-LDLb Domains are shown in green. Click for website.
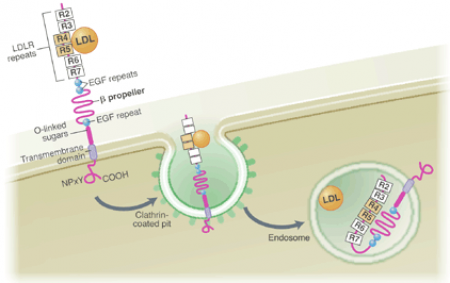
The LDLb domain, also known as LDL Receptor YWTD Domain, is critical in LDL release from LDLa domains once LDLR is internalized. YWTD is an abbreviation for the conserved residue sequence of Tyrosine-Tryptophan-Threonine-Aspartic Acid found in all LDLb domains. Both SMART and Pfam found five LDLb domains within LDLR, however it has been shown that there are actually six domains. Six repeats of LDLb domain conform into a "six-bladed" structure known as a beta-propeller (5). The beta-propeller plays an important role in LDL release from LDLa domains. Decreases in pH after internalization allows salt bridges to form between histidine residues on LDLa domains and other residues within the beta-propeller, displacing the LDL particle (1, 2). The conformational changes are specifically outlined in reference 2 and a diagram from reference 1 can be seen below. The image depicts two LDLb domains both interacting with EGF-like domains. The LDLb domains are colored green.
Analysis
SMART and Pfam were very helpful programs in order to visualize protein domains. I thought SMART provided a better representation of all domains found within LDLR, while Pfam was more useful for visualizing the 3-D structure of individual domains. By comparing figures 1 and 7, it is obvious that SMART gives a more comprehensive output. Each program provided similar information and references on the function of each domain. SMART did find the transmembrane domain responsible for anchoring LDLR into the plasma membrane, however Pfam did not. Both programs failed to find the sixth LDLb/LY domain, which is a part of the LDLR beta-propeller. This sixth domain may be difficult to find in these programs because it seems that initial domain searches only found five (5).
References
1. Innerarity, T. L. (2002). Structural biology. LDL receptor's beta-propeller displaces LDL. Science, 298(5602), 2337-9. doi: 10.1126/science.1080669
2. Rudenko, G., Henry, L., Henderson, K., Ichtchenko, K., Brown, M. S., Goldstein, J. L., Deisenhofer, J. (2002). Structure of the LDL receptor extracellular domain at endosomal pH. Science, 298(5602), 2353-8.
doi:10.1126/science.1078124
3. Appella, E., Weber, I. T., Blasi, F. (1988). Structure and function of epidermal growth factor-like regions in proteins. FEBS Lett, 231(1), 1-4. doi:10.1016/0014-5793(88)80690-2
4. Selander-Sunnerhagen, M., Ullner, M., Persson, E., Teleman, O., Stenflo, J., Drakenberg, T. (1992). How an epidermal growth factor (EGF)-like domain binds calcium. High resolution NMR structure of the calcium form of the NH2-terminal EGF-like domain in coagulation factor X. J Biol Chem, 267(27), 19642-9. PMID: 1527084
5. Springer, T. A. (1998). An extracellular beta-propeller module predicted in lipoprotein and scavenger receptors, tyrosine kinases, epidermal growth factor precursor, and extracellular matrix components. J Mol Bio, 283(4), 837-62. doi:10.1006/jmbi.1998.2115
2. Rudenko, G., Henry, L., Henderson, K., Ichtchenko, K., Brown, M. S., Goldstein, J. L., Deisenhofer, J. (2002). Structure of the LDL receptor extracellular domain at endosomal pH. Science, 298(5602), 2353-8.
doi:10.1126/science.1078124
3. Appella, E., Weber, I. T., Blasi, F. (1988). Structure and function of epidermal growth factor-like regions in proteins. FEBS Lett, 231(1), 1-4. doi:10.1016/0014-5793(88)80690-2
4. Selander-Sunnerhagen, M., Ullner, M., Persson, E., Teleman, O., Stenflo, J., Drakenberg, T. (1992). How an epidermal growth factor (EGF)-like domain binds calcium. High resolution NMR structure of the calcium form of the NH2-terminal EGF-like domain in coagulation factor X. J Biol Chem, 267(27), 19642-9. PMID: 1527084
5. Springer, T. A. (1998). An extracellular beta-propeller module predicted in lipoprotein and scavenger receptors, tyrosine kinases, epidermal growth factor precursor, and extracellular matrix components. J Mol Bio, 283(4), 837-62. doi:10.1006/jmbi.1998.2115